A Prologue to Starting a Biotech Company During the Pandemic
Our journey to develop new technology to decentralize and democratize single-cell analysis tools.
Joe de Rutte, Co-Founder at Partillion Bioscience
Dec. 2020 - Moving into our lab space in the California Nanosystems Institute.
Nearly 6 years ago I started my PhD at UCLA. Just over two years ago I graduated and started a company based on the technology I had spearheaded during that time. I wanted to take the time to reflect on this journey as (1) I have horrible memory and it would be a great idea to keep track of these things, (2) our paper on this work was finally published (I’ll let you do the math there..) and (3) potentially there are some useful nuggets of information that others can learn from.
I have broken up this post into multiple sections. Feel free to jump to the end to get the TLDR.
1. What is the problem we set out to solve?
“Our BIG advantage - no one else could see the problem we were solving”
To properly explain our story, I will need to dive into some technical jargon. Don’t worry I’ve made some great okay analogies to make it easier to follow.
I completed my PhD work in one of the world’s leading research labs in the field of microfluidics. The general premise of this field revolves around making glass chips with small (microscale) channels that can manipulate tiny volumes of fluids very precisely. This field gained a lot of interest and traction right away in the fields of molecular and cellular biology.
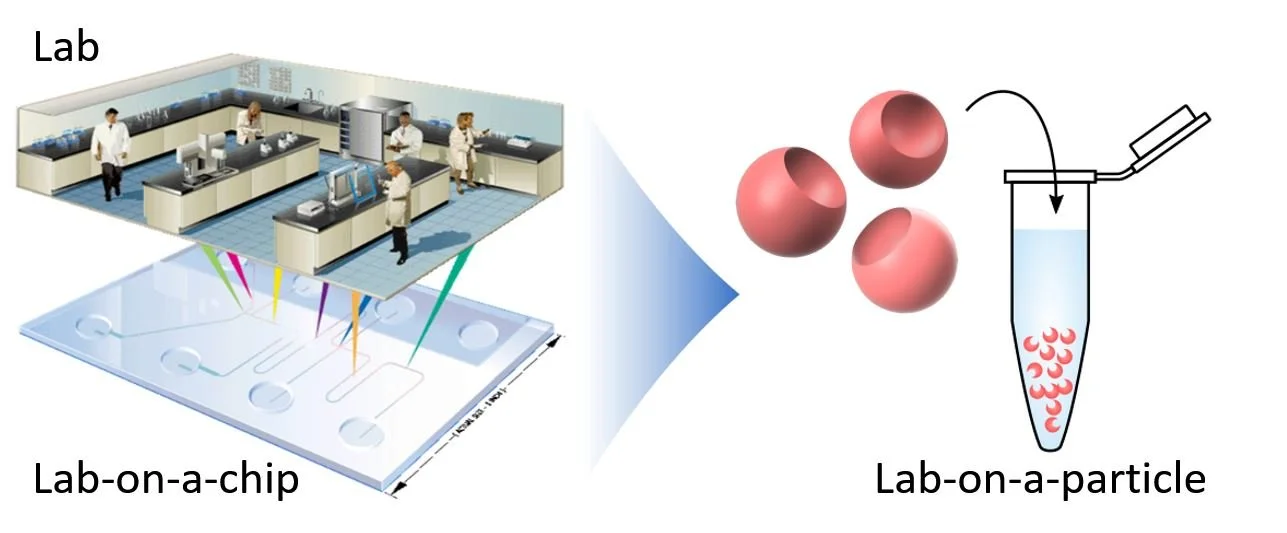
The thought was you can use this technology to miniaturize the things that you normally do in the lab and make it faster, more sensitive, cheaper, and automated. This concept was termed “Lab-on-a-chip”.
The promise of Lab-on-a-chip was to be to laboratory science as smartphones are to everyday life — do a lot of useful things in a condensed and automated format.
In concept it is great.
In reality it’s not so simple.
In contrast to computer chips — when you start messing with fluids and biology — things get really messy. Instead of making a tiny device that does everything, we often had to settle with a device that does one thing well .. aaaand requires a large complicated box of electronics, pumps, and other things for it to work.
Unlike smartphones that cost ~$1000, the cost of these boxes (“instruments) are 10’s of thousands to >$1M. On top of this the development time required can be INCREDIBLY long and take even longer for users to adopt. It is not uncommon to see new instruments take 5+ years before hitting the market.
By the time you reach the market there is often a disconnect between the upper management of companies who decides whether to acquire these fancy new toys and the researchers who are ultimately going to be using them. Colleagues at larger companies often tell stories of the expensive new toys that their company purchase only to find them sitting in a corner unused collecting dust.
Clearly this is not ideal. So what can we do better?
Many in our field would argue that the solution is simple - just build better boxes. To some extent this is a viable solution.
However, here is the problem with this approach.
A. Creating robust and easy to use instruments is REALLY difficult
This is probably one of the most underestimated aspects of developing these machines and why they are often limited to a single application — it’s HARD. The end users of these products are not technologists, they don't want spend months of effort to learn how to use the new tool only to find out it doesn't give them the data they wanted. They want something dummy proof, something that just works.
B. The cost-to-benefit ratio is often not there
It takes a lot of time and $$$ to make these instruments robust and simple, and counter to the initial promise of the field - each instrument is realistically limited to doing one or two different things. This means that to make it worth it to develop the technology in a commercial setting the market needs to be focused and it needs to be HUGE.
For some areas like single-cell sequencing this has worked great! For other areas, well you probably haven’t heard about them.
When researcher focused products did not succeed many pivoted to internal drug discovery platforms — in effect centralizing and hoarding these new technology innovations.
So how do we create a solution to this? How do we create a way of unlocking these important capabilities that microfluidics could offer, but do it in a format that is accessible to ALL researchers?
2. Proposed solution
The solution we came up with was simple in concept - what if you could do everything that these microfluidic systems are doing without actually having to use microfluidics?
Going back to the smartphone analogy, what IF, the smartphone already exists and you only need software to unlock the new features? What if we treat the lab equipment that people already have in their lab as the smartphone - the foundation, and let’s create ways to unlock new capabilities on these systems. Considering this we constructed 3 essential features that such a system would require:
Can make small compartments (the "micro" part)
Have solid surface for being able to do biochemical reactions and store information
Read out in high-throughput using existing equipment
Simple right?
When I started my PhD at UCLA and joined Prof. Di Carlo’s (Dino) lab, there were two key project areas that had been going on that were instrumental in guiding us to the solution that we had arrived at.
A. Smart biomaterials
The first revolved around using microfluidics to create micromaterials. Here the concept was using microfluidics as a way to precisely create hydrogel based microparticles that can be tuned to interact with cells in unique ways. One key use for this was a project that involved injectable materials for wound regeneration - a project that was spun out into a startup Tempo Therapeutics that is led by alumni Dr. Westbrook Weaver.
One of the key takeaways from this project was that you could use microparticles as a way to separate the microfluidics part of the work from the downstream application. We would often make a bunch of materials up front and then store them until we need to do our experiments. This also meant you don’t need the instruments used to make the materials at the same site that you are doing experiments. For instance, we had a project with a group at Texas A&M as part of the PATHS-UP engineering research center. We were able to easily create materials at one site and ship them to our collaborators to use.
B. Next generation flow cytometry
The second area was that our group had worked closely with a number of researchers developing next generation flow cytometers. Flow cytometers are instruments in which variations have existed since the 1950’s, with today’s more familiar commercial variations being released since the 1970’s. The basic operation of these devices is to readout properties of individual cells and in some cases, sort based on these properties by flowing them through a narrow region and probing them with some readout device such as a laser and sensor. These could be considered one of the first single-cell devices — decades before the term “single-cell” became a buzzword. Another astonishing fact is how ubiquitous this technology has become (>30,000 instruments worldwide) with placements at all major research institutions, cell biology labs, biotech/pharma companies.
You might now be guessing where this is going. Let’s just combine A and B.
3. Lab-on-a-chip → Lab-on-a-particle
Here was our hypothesis. What if we can create microparticles that do the microfluidic portions of our requirements:
Can make small compartments.
Have a solid surface for being able to do biochemical reactions and store information.
Be compatible with existing equipment - no new fancy instruments required.
This concept we “creatively” coined “lab-on-a-particle”.
The time it took from the point of coming up to this idea and getting to something that you could say “oh that actually works” was around 4 years.
I will not bore you with the story of all the trials and tribulations but will give you some highlights of insights we got along the way.
Keep things simple — if you think it’s already simple, think again
There have been a few aha! moments that I remember distinctly — usually when brainstorming with my advisor Dino in his office during a weekly check in. These moments usually started with the simple questions of "Are we over thinking this?", "Could we just do this instead?" The answer was usually yes, but sometimes we didn’t recognize it right away. Often with your nose deep into research it’s hard to take a step back and question your foundational assumptions. But when you do take a step back and get some fresh air, those are often the times when you make the biggest leaps forward.
We have adopted this concept of reductive problem solving as one of our key values at Partillion. There is a great article about this concept linked below.
Innovation grounded by first principles thinking
My PhD advisor and co-founder Dino Di Carlo is a researcher that has faced his fair share of skepticism — typically at the onset of new ideas (some of which later became subcategories of our field). You wouldn’t think that now given how well known and respected he is in our field. If you ask him today, Dino will laugh and tell you that people are still skeptical.
Psychologically I think most people tend to gravitate towards following the paths set by others before them. They draw on what people have done and what they are currently doing and if it falls outside of that box, they dismiss it because it doesn't fall within their current framework. This is only natural as it is a very fast and efficient way to learn and adapt to our environment. However, as scientists and engineers who are trying to push the boundaries of what is known and what is possible, we should only dismiss ideas with solid reasoning, not reactionary skepticism.
To innovate you need to look outside of the box while sticking to fundamental principles.
A great quote from Dino that I remind myself of quite often when receiving skepticism is
“Is there any good reason not to do this or why this wouldn’t work?”
If the answer is no, then there is no reason not to try. Simple as that.
As we kicked off this project we received our share of feedback, plenty of which was telling us “politely” that our idea is dumb. There is a folder on my computer where I keep these gems.
One consistent line of feedback that I think is the most interesting was some variation of
“Oh, this is interesting, but it doesn’t seem to be doing anything new, aren’t there already microfluidic approaches for doing this?” See below for an actual exert from a grant review from 2018.
“The proposed project reflects PI’s vision that creation of such 3D drop-carrier particles is a central technology for single cell biology. Although the reviewer does not necessarily share the same vision as the PI, the technology is indeed innovative and offers an interesting option that may facilitate certain single cell analyses. However, it does not appear to play the central role of transforming biological research."
The comment doesn’t actually provide logical reasoning beyond just stating I don't believe this is important.
At first this was really frustrating. Why do people not see this vision or recognize why it's even important?
Looking back now this was in many ways a HUGE advantage. We had an insight to the problem that most others were not aware of. In part due to how deep into the field we were - note that Dino has gone through the process of starting multiple companies revolving around commercializing microfluidic technologies and had firsthand experience on why this process was so painful — and that we had taken the step back to re-imagine alternative paths.
4. Turning it into reality — confirming our hypothesis
Ultimately, we came up with a solution that ticked all our required boxes. The solution we refer to now as "nanovials" are hydrogel particles with a cavity that acts as a container for holding individual cells.
Here is a 3D printed version we like to show people as for some reason it’s easier to understand.
Image: (Left) microscopy image of nanovials with cells inside and fluorescent signal from secreted proteins. (Right) 3D Printed Nanovial with a coffee bean inside to represent a cell.
The first application space we focused on was using the technology for being able to analyze and sort individual cells based on the proteins they are secreting. This is critical from a fundamental biology perspective — >1900 proteins are secreted (Protein Atlas) — and is important in the discovery and development of some of the most cutting-edge therapeutics such as monoclonal antibodies and cell therapies. Note, 5/10 of the blockbuster drugs in 2019 were monoclonal antibodies (Pharma Intelligence), and cell based therapies continue to see year to year growth, with over 2000 active clinical trials in 2021, up 38% from 2020 (Nature Reviews Drug Discovery).
When we showcased our first demonstration of our technology there was a notable shift in people’s reception of the idea. In our own field we received positive feedback from other researchers who could now better understand the motivation and see the potential. More notably there was incredibly positive reception at more industry focused events. This was eye opening to us as it was very clear that those who would potentially benefit the most from the technology recognized it as a solution. In February 2020 I received the top $10,000 innovation award for this work at the Society of Laboratory Automation and Screening.
It was after this event , and another event that happened around that same time that motivated us to make the jump to start Partillion Bioscience which I now lead and we recently launched our first commercial product.
In parallel to this we started submitting the first version of our paper for publication back in April of 2020. The reception that we got from academic journals did not at all correlate with the reception we got from researchers on the industry side. Perhaps another indicator of the frequently large disconnect between academic research and commercial implementation — academic journals incentivize and promote work that is on the “bleeding” edge which often translates to being incredibly difficult and, in many cases, not replicable leading many people astray (Serra-Garcia,& Gneezy, Sci. Adv. 2021).
So what did we do? Well we posted our paper on bioRxiv and kept grinding at the research to try and improve the manuscript. It took two years before we finally published the work, eventually finding a home in ACS Nano. Funny enough we had literally started, and published two follow-up papers within that same time period. For those outside of academia this might sound crazy, for those that are in academia it’s familiar.
One interesting thing that we noticed during this whole time is that no one really cared that the paper was a preprint. People who understood the technology and space didn’t need some fancy journal to sign off on it to understand it was important.
In total we only spent one more year collecting additional data for the paper. When we first submitted the paper there were 5 authors; when we finally published the paper there were 19 authors. Why do I highlight this? I see the growth in those working on this project as a testament to what the technology set out to do; create an accessible approach that anyone could use anywhere. For instance, we worked with Spangler lab based at Johns Hopkins to perform experiments on antibody producing cell lines that they had. We were able to send them materials and have them run the experiments completely remotely on the equipment they already had. It was also a testament to how excited people were to work on the project and saw the potential of it. Many of these early collaborators are continuing to use the technology today to gain new insights in to their research. Within that first 2 years (right after the pandemic) we were able to seed over 20 collaborations with various research labs and commercial partners - nearly all of which are completely remote.
5. Closing thoughts
General take-Aways
Building deep knowledge of a field gives you opportunity to unearth hidden problems to solve.
If you find a hidden problem you want to solve – be prepared to receive a lot of skepticism.
Don’t focus on what haters say, focus on their reasoning.
Creating a great solution to problems can take years and many iterations.
Keep in mind the bigger goal and be flexible on what path gets you there.
Have fun along the way – life’s short.
Outlook on microfluidics and single-cell technology moving forward.
Moving forward we see the microfluidic field splitting in two directions. We will still see companies and research groups continue to build complex systems that will be used for services or for internal discovery pipelines. This was observed recently as companies such as BLI have shifted more and more towards partnership/service-based business models (BLI Press Release) or companies such as AbCellera that focused on this from the beginning. In some cases we see companies shifting even further away with one of the first microfluidic companies Fluidigm recently rebranded to Standard BioTools after receiving a $250M capital reinfusion, noting a focus towards its mass cytometry based products (GlobalNewsWire).
On the other side I think we will see a larger shift towards creating simplified solutions that escape the standard make a box model. The key here will be leveraging the infrastructure that is already in place. One of the earliest examples of this is how Luminex leveraged a large install base of flow cytometers for its multi-analyte profiling technology back in the late 90’s. Even more recently companies such as 10X Genomics were in part able to grow so quickly by leveraging the large install base of Illumina’s next-gen sequencing technology as a final readout. They still have a box, but its relatively cheap compared to the consumable costs reducing it as a barrier to adoption.
We expect this to go even further — completely removing the box and creating even simpler and more flexible systems that empower researchers. Microfluidics does not necessarily equal complex microchips.
This is where we put our bets down 6 years ago and where we will continue to double down.
Disclaimer: Joe de Rutte is Co-founder and president of Partillion Bioscience which commercializes next generation single-cell technology.